Volume 11, Issue 1 (Winter 2023)
Iran J Health Sci 2023, 11(1): 47-58 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Jaryani F, Amiri M. A Pre-Trained Ensemble Model for Breast Cancer Grade Detection Based on Small Datasets. Iran J Health Sci 2023; 11 (1) :47-58
URL: http://jhs.mazums.ac.ir/article-1-816-en.html
URL: http://jhs.mazums.ac.ir/article-1-816-en.html
Department of Computer Engineering, Faculty of Engineering, Arak University, Arak, Iran. , m-amiri@araku.ac.ir
Full-Text [PDF 2805 kb]
(1040 Downloads)
| Abstract (HTML) (2210 Views)
Full-Text: (1011 Views)
1. Introduction
Cancer is one of the significant global public health issues and the second leading cause of death [1]. The institute for health metrics and evaluation (IHME) reports that cancer caused an estimated 9.6 million deaths in 2017, as shown in Figure 1 [2].
.jpg)
Figure 2 shows the total number of deaths caused by various cancers across all ages and genders [3]. The leading reason for death is stomach cancer, while breast cancer ranks as the fifth cause of death.
.jpg)
Invasive ductal carcinoma (IDC) is recognized as one of the most widespread phenotypic sub-classification of all breast cancers since nearly 80% of identified breast cancers are IDC [4]. It develops in milk ducts and conquers the fibrous tissue of the breast outside the duct. As Figure 3 shows, pathologists recognize the cancer type and grade through visual investigation of tissue stained by hematoxylin and eosin (H&E) [5].

The precise detection of IDC grade has a significant effect on determining a proper treatment plan. The histologic grade is a prognostic factor and an indicator of response to chemotherapy. It is closely related to the frequency of recurrence and death due to IDC, disease-free interval, and longer life after mastectomy. Several studies show that patients with a high-grade IDC treated with a mastectomy had a remarkably higher frequency of auxiliary lymph node (ALN) and mortality rate than the patients with a lower-grade IDC. The high-grade carcinomas result in early treatment failures, while subsequent recurrences are more often observed among low-grade tumors [6, 7].
The emphasis on early detection of breast cancer is due to the significant impact on the survival rate. According to acute coronary syndrome (ACS), the 5-year survival rate for localized breast cancer (cancer that has not spread beyond the breast) is 99%. For patients with regional breast cancer (cancer that has spread to nearby lymph nodes), that number falls to 86%; for cancer that has spread to more distant parts of the body, the 5-year survival rate is only 29%.
The latest proposed breast cancer detectors are based on machine learning (ML) techniques, such as deep learning (DL). DL is a form of ML that can utilize supervised [4, 8], or unsupervised algorithms [8, 9, 10]. DL supports multi-layer hybrid models to gather data [11]. AlexNet [12], VGG net [13], ResNet [7], ResNeXt [14], and RCNN (region-based convolutional neural network) [15, 16, 17] are considered as DL’s advanced models. Most previous works in the field of cancer detection focus on large datasets and are not developed for small datasets. Although large datasets may lead to more reliable results, their collecting and processing are challenging. This paper proposes a new ensemble deep-learning model for breast cancer grade detection based on small datasets. Our model uses MobileNetV2, VGG16, and EfficientNet-B0 as pretrained basic transfer learning classifiers to grade the breast tumors, including grades I, II, and III. The features learned from the deep models make the deep network very effective in problems with small-sized datasets.
The rest of the paper is organized as follows, section 2 reviews related works on breast cancer diagnosis. Section 3 introduces the proposed ensemble model. We present the experimental results in section 4. Section 5 indicates the discussion, and finally, the paper concludes with our future work in section 6.
Related Works
Recently, various efforts have been conducted to detect and protect various types of cancer by applying different artificial intelligence methods [13, 18, 19, 20]. The effective usage of multiple data features and the assortment of acceptable classification techniques [19, 21] is significant for image classification. Table 1 summarizes the previous breast cancer diagnosis models based on ML.
.jpg)
As the table shows, most works focus on large datasets. Moreover, augmentation techniques have not been used in previous models. In this paper, we propose a new ensemble model for breast cancer detection focusing on small datasets and using augmentation techniques.
2. Materials and Methods
Proposed Model
In this paper, we use ImageNet as a pre-trained model on a large-scale dataset. The model is modified to distinguish different images. The trained features of deep models can lead to a deep network that solves problems with limited-size datasets. The VGG16, Inception V3, and Inception-ResNetV2 models are applied to small-sized breast cancer datasets. The EfficientNet-B0 model is considered a standard model for image classification of the limited-size dataset with acceptable performance. Weights trained on ImageNet are used for VGG16, Inception V3, and ResNet 50 deep models. For the baseline, EfficientNet-B0 is rum directly with the binary classifier as an optimizer without any data dropout or augmentation.
While the training time of the pre-trained networks is relatively low, their accuracy is high. Therefore, the use of pre-trained deep learning methods can increase prediction accuracy and improve image classification. Furthermore, applying well-tuned ensemble models can significantly improve the model’s performance. In the following subsections, the proposed model is considered in detail.
Dataset
In this paper, a new dataset, Databiox [4], is used. The Databiox dataset has been chosen to grade breast cancers in three different classes named grades I, II, and III. Databiox is a dataset for histopathological microscopy images of patients with IDC containing 922 images. All images are in RGB format with JPEG type. The resolution of the images is 2100 × 1574 or 1276 × 956 pixels. Table 2 presents the technical specifications of the dataset in detail.
.jpg)
The specimens are breast tissues stained with H&E, obtained from 124 patients diagnosed between 2014 and 2019 in the PourSina Hakim research center of Isfahan University of Medical Sciences in Iran. Each specimen is provided with four magnification levels: 4x,10x, 20x, and 40x as shown in Figure 4.

The labeling of all images in the dataset based on their diagnosed grade can be used to train machine learning algorithms to recognize IDC grades. This dataset uses the same grading method as previous datasets but differs from others in that it contains an equal number of samples from each of the three IDC grades, resulting in approximately 50 samples for each grade [28].
Based on the pathologist’s opinion, more than one image from a specific magnification level is presented in some cases. For example, four 40x images exist for almost all of the specimens. A score of 1, 2, or 3 is assigned to each, and scores are added to produce a grade. Table 3 presents how the Databiox images have been categorized in each grade and zoom.
.jpg)
Dataset modification
According to section 2, none of the previous works provide breast cancer classification based on the grades. In this paper, breast cancer classification is performed on the Databiox dataset, which is based on the grades. As mentioned, this paper aims to work on small datasets, such as Databiox. For small datasets, it seems a hierarchical augmentation method can generate a larger dataset that is more reliable for the testing and validation process. To generate an augmented image, a series of pre-processing transformations, including horizontal and vertical flipping, skewing, cropping, rotating, and zooming is performed on the original image. Indeed, the augmented images can reproduce different data points. It is opposed to just duplicating the same data. The subtle differences of these “additional” images should be enough to train a more robust model. Two horizontal and vertical flipping are employed to zoom the dataset images from 80% to 100%. Figure 5 shows the augmentation methods for a sample image selected from Databiox with both the flipping and zooming augmentation applied to this sample image. The total number of images after applying the augmentation methods is .

The success of image classification depends on the quality and quantity of the training dataset. With a larger dataset, we will have a more effective deep-learning model for image processing. Data augmentation increases the size and diversity of datasets without manually collecting new images.
3. Results
Proposed ensemble model
The goal of the proposed model is to create an efficient ensemble trainer for the diagnosis of breast cancer grades I, II, and III. Ensemble methods employ several machine learning algorithms to reach acceptable performance more than each of the individual methods. Figure 6 shows the main steps of the proposed ensemble model. The steps are explained in the following subsections.

Step 1: Training individual models and saving them
First, all individual models are created by applying three different models MobileNetV2, VGG16, and efficientNEtb0. Then, the weights are loaded, it chooses to freeze or unfreeze loaded weights, and finally, a dens layer is added to the outputs. In the next step, the accuracy and loss of the individual models are compared. Figure 7 illustrates accuracy and loss in the training and validation phases for MobileNetv2 in 20 training epochs.
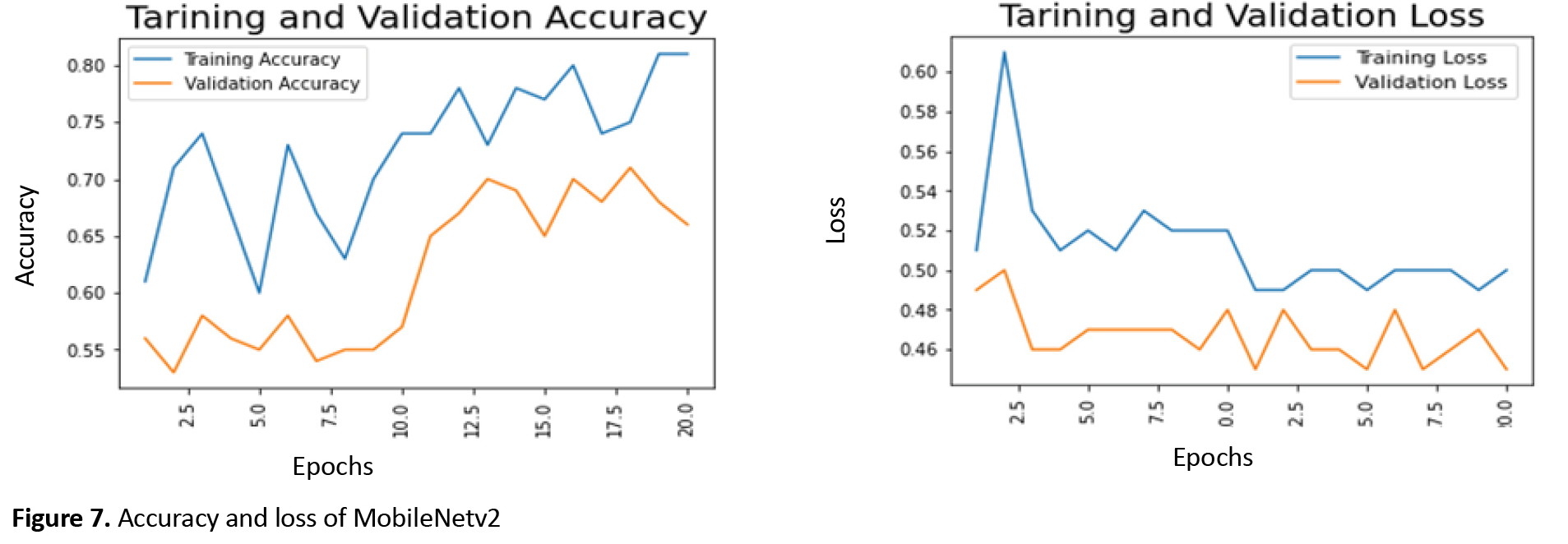
The maximum training accuracy for MobileNetv2 is around 81%, and the minimum training loss is about 50%, which occurs after 20 epochs. Additionally, the best validation accuracy after 20 epochs is about 70%, which can be seen in epoch 18.
In a similar way to MobileNetv2, Figure 8 shows training and validation loss and accuracy for efficinetNetB0 in 20 training epochs.

The best training accuracy for efficinetNetB0 is around 77%, and the minimum training loss is 50%, which occurs after 20 epochs. Moreover, the maximum validation accuracy after 20 epochs is about 70% Figure 9 shows the accuracy and loss of the training and validation for VGG16 in 20 training epochs.
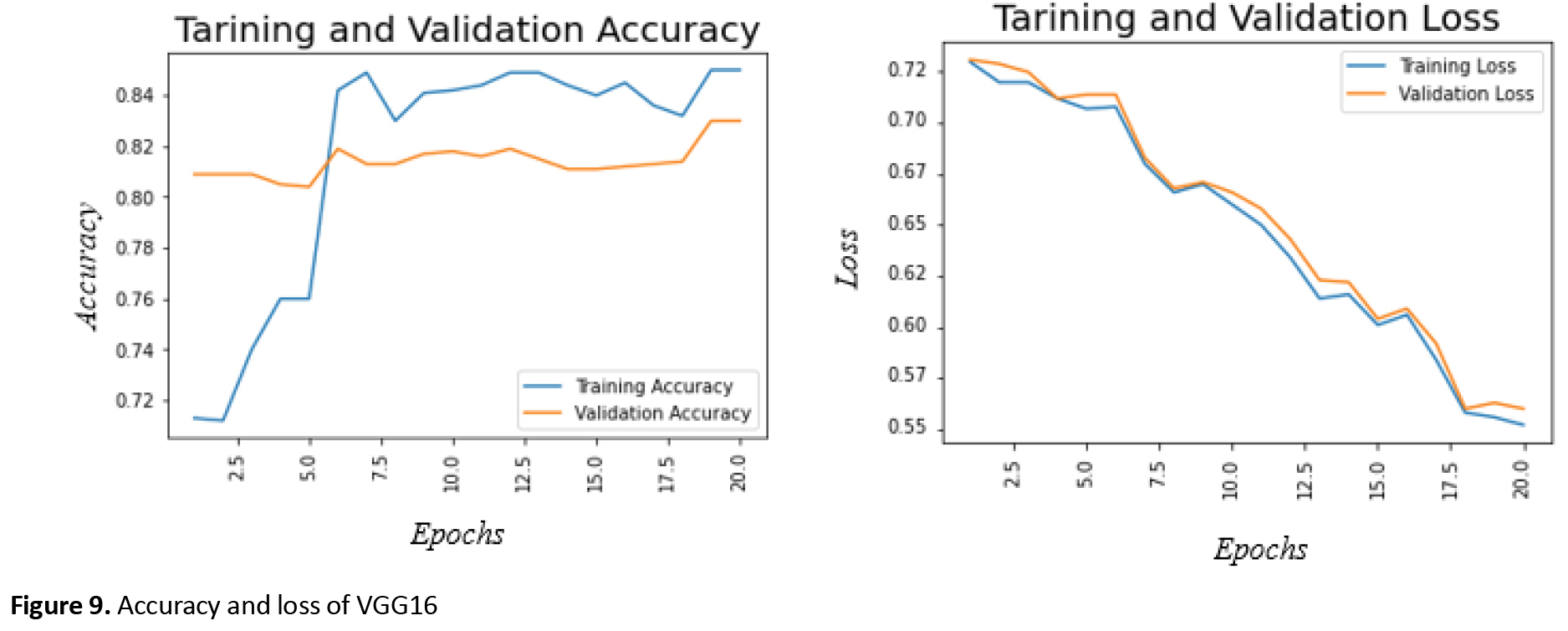
The maximum training accuracy for VGG16 is 76%, and the minimum training loss is 50%, which occurs after 20 epochs. In addition, the best validation accuracy after 20 epochs is 57%.
Steps 2 to 4: The final ensemble model
Figure 10 shows the proposed ensemble framework for breast cancer classification. The framework consists of three different classification models, including EfficientNet-B0, MobileNet v2, and VGG16. EfficientNet-B0 is a new scaling method called hybrid scaling. It suggests that we will get much better performance if we simultaneously scale the dimensions by a fixed amount and do it uniformly. The scaling coefficients can be set by the user. The VGG16 is one of the most popular pre-trained models for image classification. VGG-16 broke the AlexNet standards and was quickly adopted by researchers and the industry for their image classification tasks. The MobileNet-v2 architecture is based on an inverted residual structure where the input and output of the residual block are thin bottleneck layers opposite to traditional residual models that use expanded representations in the input. MobileNet v2 uses lightweight depth-wise convolutions to filter features in the intermediate expansion layer.

The individual models and their freeze layers are loaded in the next step so that their weight does not change when the ensemble model is fitted on them. Then we concatenate their outputs and add dense layers. As shown in Figure 10, the proposed model takes the outputs of all the models and puts them in a concatenated layer. Since the proposed method is a multiclass classifier, a dense layer with a sigmoid activation function is used. This procedure is similar to neural networks, where the predictions of all the models are taken as inputs and an output is provided.
Evaluation
To evaluate the proposed model, both individual models and the ensemble models are trained for 20 epochs. Figure 11 shows the accuracy and loss of the ensemble model in the training and validation phases. Table 4 presents the validation accuracy of the models on their final epochs.

.jpg)
As the Table shows, the ensemble model improves validation accuracy compared to the individual models. In other words, data augmentation and powerful dropout on a pre-trained deep model lead to very high accuracy. Furthermore, it shows that small datasets can also use the power of depth. Creating an ensemble model is a very long procedure that requires putting more effort and perseverance into a model. However, achieving this accuracy would be valuable, especially utilizing small datasets that were initially unlikely.
The accuracy of the proposed ensemble model is increased by almost 11%, which is remarkable considering that the previous best accuracy was 70% obtained with EfficientNet-B0. However, building an ensemble model in this way is a time-consuming process. It is three times the effort of a single model but can contribute to accuracy that is difficult to achieve in small data sets.
4. Discussion
Few similar studies have been conducted in this area, e.g., Faisal et al. in [29] used the technology of “gradient boosting tree (GBT) with majority voting and radio frequency (RF)-based ensembles” and its accuracy rate was 90%. Bhowal et al. in [30] also used the technology “Choquet integral-based deep CNN models with coalition game and information theory”. They achieved 95% accuracy in determining breast cancer grade. These two models show better accuracy than our proposed model because they use large datasets for training. In contrast, our dataset is a small dataset collected from Iranian patients.
Few works train the entire model from the beginning since a significant dataset is rarely provided. Moreover, for feature extraction, most researchers are interested in using a model pre-trained on a widespread dataset, such as ImageNet. Subsequently, researchers introduce transfer learning to optimize time and achieve performance improvement simultaneously.
Recently, several considerable attempts have been made to detect and predict all types of cancer. Artificial intelligence (AI) and its subdomains including machine learning and deep learning are widely used for breast cancer detection. Reviewing several related works conducted in recent years [10, 11, 12, 13, 14] validates the importance of machine learning (ML) methods for the classification of the histopathological microscopic image dataset to upgrade IDC. Hence, an ensemble model is proposed for image classification on the Databiox dataset. Table 5 compares our proposed model with the above-tested deep learning models.
.jpg)
The critical limitation of this study occurs when a transformed sample differs significantly from the original sample in terms of pixels. So, the network may not be able to classify it correctly. However, we can train the model for a particular transformation on the transformed data to achieve high accuracy.
An important question is whether deep models, such as region-based CNN (RCNN) [14], and you only look once (YOLO) [31] can be fitted to a very small dataset or not. We plan to qualify more models on very small datasets and compare their performance in our future work.
5. Conclusion
In this study, Databiox, a new small dataset, was used to grade breast cancer in three different grades I, II, and III. Since the Databiox is very small and only includes images taken from 124 patients with a total of 924 images, we have applied the augmentation techniques to extend the dataset and consequently improve the accuracy of the proposed methods.
The proposed ensemble model is constructed based on the single models MobileNetV2, VGG16, and EfficientNet-B0Seven, and its performance is compared to these single models. In this paper, we pre-trained the proposed model based on large-scale datasets and applied these qualified models to small-size datasets with great performance. The experiments confirm that the ensemble model can be used for small datasets with precise modifications by applying the proper augmentation techniques. In addition, the findings indicate that the proposed method achieves the best accuracy among all the models. This achievement is essential to design future classification-based systems in computer-aided diagnosis since it shows that the use of ensemble models can improve the accuracy of breast cancer diagnosis, especially for small datasets.
In future work, we work on increasing the classification accuracy. In addition, we plan to consider the impact of the other single deep models on the performance of the ensemble model on small datasets and lower-resolution images.
Availability of Data and Materials:
The dataset used in this study can be found at http://databiox.com/datasets. The codes can be provided, upon reasonable request. They may be available from the corresponding author.
Ethical Considerations
Compliance with ethical guidelines
There were no ethical considerations to be considered in this research.
Funding
This research was supported by Arak University (Grant No.: 98/11402).
Authors contributions
Proposing the model and developing the proposed model, drafting and editing the manuscript, drafting and editing the figures: Farhang Jaryani; Initiating and coordinating the project, interpreting the data, editing the manuscript, and editing the figures: Maryam Amiri.
Conflict of interest
The authors declared no conflict of interest.
Acknowledgements
We thank Iran’s National Elites Foundation and Arak University for their support in funding this research.
References
Cancer is one of the significant global public health issues and the second leading cause of death [1]. The institute for health metrics and evaluation (IHME) reports that cancer caused an estimated 9.6 million deaths in 2017, as shown in Figure 1 [2].
.jpg)
Figure 2 shows the total number of deaths caused by various cancers across all ages and genders [3]. The leading reason for death is stomach cancer, while breast cancer ranks as the fifth cause of death.
.jpg)
Invasive ductal carcinoma (IDC) is recognized as one of the most widespread phenotypic sub-classification of all breast cancers since nearly 80% of identified breast cancers are IDC [4]. It develops in milk ducts and conquers the fibrous tissue of the breast outside the duct. As Figure 3 shows, pathologists recognize the cancer type and grade through visual investigation of tissue stained by hematoxylin and eosin (H&E) [5].

The precise detection of IDC grade has a significant effect on determining a proper treatment plan. The histologic grade is a prognostic factor and an indicator of response to chemotherapy. It is closely related to the frequency of recurrence and death due to IDC, disease-free interval, and longer life after mastectomy. Several studies show that patients with a high-grade IDC treated with a mastectomy had a remarkably higher frequency of auxiliary lymph node (ALN) and mortality rate than the patients with a lower-grade IDC. The high-grade carcinomas result in early treatment failures, while subsequent recurrences are more often observed among low-grade tumors [6, 7].
The emphasis on early detection of breast cancer is due to the significant impact on the survival rate. According to acute coronary syndrome (ACS), the 5-year survival rate for localized breast cancer (cancer that has not spread beyond the breast) is 99%. For patients with regional breast cancer (cancer that has spread to nearby lymph nodes), that number falls to 86%; for cancer that has spread to more distant parts of the body, the 5-year survival rate is only 29%.
The latest proposed breast cancer detectors are based on machine learning (ML) techniques, such as deep learning (DL). DL is a form of ML that can utilize supervised [4, 8], or unsupervised algorithms [8, 9, 10]. DL supports multi-layer hybrid models to gather data [11]. AlexNet [12], VGG net [13], ResNet [7], ResNeXt [14], and RCNN (region-based convolutional neural network) [15, 16, 17] are considered as DL’s advanced models. Most previous works in the field of cancer detection focus on large datasets and are not developed for small datasets. Although large datasets may lead to more reliable results, their collecting and processing are challenging. This paper proposes a new ensemble deep-learning model for breast cancer grade detection based on small datasets. Our model uses MobileNetV2, VGG16, and EfficientNet-B0 as pretrained basic transfer learning classifiers to grade the breast tumors, including grades I, II, and III. The features learned from the deep models make the deep network very effective in problems with small-sized datasets.
The rest of the paper is organized as follows, section 2 reviews related works on breast cancer diagnosis. Section 3 introduces the proposed ensemble model. We present the experimental results in section 4. Section 5 indicates the discussion, and finally, the paper concludes with our future work in section 6.
Related Works
Recently, various efforts have been conducted to detect and protect various types of cancer by applying different artificial intelligence methods [13, 18, 19, 20]. The effective usage of multiple data features and the assortment of acceptable classification techniques [19, 21] is significant for image classification. Table 1 summarizes the previous breast cancer diagnosis models based on ML.
.jpg)
As the table shows, most works focus on large datasets. Moreover, augmentation techniques have not been used in previous models. In this paper, we propose a new ensemble model for breast cancer detection focusing on small datasets and using augmentation techniques.
2. Materials and Methods
Proposed Model
In this paper, we use ImageNet as a pre-trained model on a large-scale dataset. The model is modified to distinguish different images. The trained features of deep models can lead to a deep network that solves problems with limited-size datasets. The VGG16, Inception V3, and Inception-ResNetV2 models are applied to small-sized breast cancer datasets. The EfficientNet-B0 model is considered a standard model for image classification of the limited-size dataset with acceptable performance. Weights trained on ImageNet are used for VGG16, Inception V3, and ResNet 50 deep models. For the baseline, EfficientNet-B0 is rum directly with the binary classifier as an optimizer without any data dropout or augmentation.
While the training time of the pre-trained networks is relatively low, their accuracy is high. Therefore, the use of pre-trained deep learning methods can increase prediction accuracy and improve image classification. Furthermore, applying well-tuned ensemble models can significantly improve the model’s performance. In the following subsections, the proposed model is considered in detail.
Dataset
In this paper, a new dataset, Databiox [4], is used. The Databiox dataset has been chosen to grade breast cancers in three different classes named grades I, II, and III. Databiox is a dataset for histopathological microscopy images of patients with IDC containing 922 images. All images are in RGB format with JPEG type. The resolution of the images is 2100 × 1574 or 1276 × 956 pixels. Table 2 presents the technical specifications of the dataset in detail.
.jpg)
The specimens are breast tissues stained with H&E, obtained from 124 patients diagnosed between 2014 and 2019 in the PourSina Hakim research center of Isfahan University of Medical Sciences in Iran. Each specimen is provided with four magnification levels: 4x,10x, 20x, and 40x as shown in Figure 4.

The labeling of all images in the dataset based on their diagnosed grade can be used to train machine learning algorithms to recognize IDC grades. This dataset uses the same grading method as previous datasets but differs from others in that it contains an equal number of samples from each of the three IDC grades, resulting in approximately 50 samples for each grade [28].
Based on the pathologist’s opinion, more than one image from a specific magnification level is presented in some cases. For example, four 40x images exist for almost all of the specimens. A score of 1, 2, or 3 is assigned to each, and scores are added to produce a grade. Table 3 presents how the Databiox images have been categorized in each grade and zoom.
.jpg)
Dataset modification
According to section 2, none of the previous works provide breast cancer classification based on the grades. In this paper, breast cancer classification is performed on the Databiox dataset, which is based on the grades. As mentioned, this paper aims to work on small datasets, such as Databiox. For small datasets, it seems a hierarchical augmentation method can generate a larger dataset that is more reliable for the testing and validation process. To generate an augmented image, a series of pre-processing transformations, including horizontal and vertical flipping, skewing, cropping, rotating, and zooming is performed on the original image. Indeed, the augmented images can reproduce different data points. It is opposed to just duplicating the same data. The subtle differences of these “additional” images should be enough to train a more robust model. Two horizontal and vertical flipping are employed to zoom the dataset images from 80% to 100%. Figure 5 shows the augmentation methods for a sample image selected from Databiox with both the flipping and zooming augmentation applied to this sample image. The total number of images after applying the augmentation methods is .

The success of image classification depends on the quality and quantity of the training dataset. With a larger dataset, we will have a more effective deep-learning model for image processing. Data augmentation increases the size and diversity of datasets without manually collecting new images.
3. Results
Proposed ensemble model
The goal of the proposed model is to create an efficient ensemble trainer for the diagnosis of breast cancer grades I, II, and III. Ensemble methods employ several machine learning algorithms to reach acceptable performance more than each of the individual methods. Figure 6 shows the main steps of the proposed ensemble model. The steps are explained in the following subsections.

Step 1: Training individual models and saving them
First, all individual models are created by applying three different models MobileNetV2, VGG16, and efficientNEtb0. Then, the weights are loaded, it chooses to freeze or unfreeze loaded weights, and finally, a dens layer is added to the outputs. In the next step, the accuracy and loss of the individual models are compared. Figure 7 illustrates accuracy and loss in the training and validation phases for MobileNetv2 in 20 training epochs.

The maximum training accuracy for MobileNetv2 is around 81%, and the minimum training loss is about 50%, which occurs after 20 epochs. Additionally, the best validation accuracy after 20 epochs is about 70%, which can be seen in epoch 18.
In a similar way to MobileNetv2, Figure 8 shows training and validation loss and accuracy for efficinetNetB0 in 20 training epochs.

The best training accuracy for efficinetNetB0 is around 77%, and the minimum training loss is 50%, which occurs after 20 epochs. Moreover, the maximum validation accuracy after 20 epochs is about 70% Figure 9 shows the accuracy and loss of the training and validation for VGG16 in 20 training epochs.

The maximum training accuracy for VGG16 is 76%, and the minimum training loss is 50%, which occurs after 20 epochs. In addition, the best validation accuracy after 20 epochs is 57%.
Steps 2 to 4: The final ensemble model
Figure 10 shows the proposed ensemble framework for breast cancer classification. The framework consists of three different classification models, including EfficientNet-B0, MobileNet v2, and VGG16. EfficientNet-B0 is a new scaling method called hybrid scaling. It suggests that we will get much better performance if we simultaneously scale the dimensions by a fixed amount and do it uniformly. The scaling coefficients can be set by the user. The VGG16 is one of the most popular pre-trained models for image classification. VGG-16 broke the AlexNet standards and was quickly adopted by researchers and the industry for their image classification tasks. The MobileNet-v2 architecture is based on an inverted residual structure where the input and output of the residual block are thin bottleneck layers opposite to traditional residual models that use expanded representations in the input. MobileNet v2 uses lightweight depth-wise convolutions to filter features in the intermediate expansion layer.

The individual models and their freeze layers are loaded in the next step so that their weight does not change when the ensemble model is fitted on them. Then we concatenate their outputs and add dense layers. As shown in Figure 10, the proposed model takes the outputs of all the models and puts them in a concatenated layer. Since the proposed method is a multiclass classifier, a dense layer with a sigmoid activation function is used. This procedure is similar to neural networks, where the predictions of all the models are taken as inputs and an output is provided.
Evaluation
To evaluate the proposed model, both individual models and the ensemble models are trained for 20 epochs. Figure 11 shows the accuracy and loss of the ensemble model in the training and validation phases. Table 4 presents the validation accuracy of the models on their final epochs.

.jpg)
As the Table shows, the ensemble model improves validation accuracy compared to the individual models. In other words, data augmentation and powerful dropout on a pre-trained deep model lead to very high accuracy. Furthermore, it shows that small datasets can also use the power of depth. Creating an ensemble model is a very long procedure that requires putting more effort and perseverance into a model. However, achieving this accuracy would be valuable, especially utilizing small datasets that were initially unlikely.
The accuracy of the proposed ensemble model is increased by almost 11%, which is remarkable considering that the previous best accuracy was 70% obtained with EfficientNet-B0. However, building an ensemble model in this way is a time-consuming process. It is three times the effort of a single model but can contribute to accuracy that is difficult to achieve in small data sets.
4. Discussion
Few similar studies have been conducted in this area, e.g., Faisal et al. in [29] used the technology of “gradient boosting tree (GBT) with majority voting and radio frequency (RF)-based ensembles” and its accuracy rate was 90%. Bhowal et al. in [30] also used the technology “Choquet integral-based deep CNN models with coalition game and information theory”. They achieved 95% accuracy in determining breast cancer grade. These two models show better accuracy than our proposed model because they use large datasets for training. In contrast, our dataset is a small dataset collected from Iranian patients.
Few works train the entire model from the beginning since a significant dataset is rarely provided. Moreover, for feature extraction, most researchers are interested in using a model pre-trained on a widespread dataset, such as ImageNet. Subsequently, researchers introduce transfer learning to optimize time and achieve performance improvement simultaneously.
Recently, several considerable attempts have been made to detect and predict all types of cancer. Artificial intelligence (AI) and its subdomains including machine learning and deep learning are widely used for breast cancer detection. Reviewing several related works conducted in recent years [10, 11, 12, 13, 14] validates the importance of machine learning (ML) methods for the classification of the histopathological microscopic image dataset to upgrade IDC. Hence, an ensemble model is proposed for image classification on the Databiox dataset. Table 5 compares our proposed model with the above-tested deep learning models.
.jpg)
The critical limitation of this study occurs when a transformed sample differs significantly from the original sample in terms of pixels. So, the network may not be able to classify it correctly. However, we can train the model for a particular transformation on the transformed data to achieve high accuracy.
An important question is whether deep models, such as region-based CNN (RCNN) [14], and you only look once (YOLO) [31] can be fitted to a very small dataset or not. We plan to qualify more models on very small datasets and compare their performance in our future work.
5. Conclusion
In this study, Databiox, a new small dataset, was used to grade breast cancer in three different grades I, II, and III. Since the Databiox is very small and only includes images taken from 124 patients with a total of 924 images, we have applied the augmentation techniques to extend the dataset and consequently improve the accuracy of the proposed methods.
The proposed ensemble model is constructed based on the single models MobileNetV2, VGG16, and EfficientNet-B0Seven, and its performance is compared to these single models. In this paper, we pre-trained the proposed model based on large-scale datasets and applied these qualified models to small-size datasets with great performance. The experiments confirm that the ensemble model can be used for small datasets with precise modifications by applying the proper augmentation techniques. In addition, the findings indicate that the proposed method achieves the best accuracy among all the models. This achievement is essential to design future classification-based systems in computer-aided diagnosis since it shows that the use of ensemble models can improve the accuracy of breast cancer diagnosis, especially for small datasets.
In future work, we work on increasing the classification accuracy. In addition, we plan to consider the impact of the other single deep models on the performance of the ensemble model on small datasets and lower-resolution images.
Availability of Data and Materials:
The dataset used in this study can be found at http://databiox.com/datasets. The codes can be provided, upon reasonable request. They may be available from the corresponding author.
Ethical Considerations
Compliance with ethical guidelines
There were no ethical considerations to be considered in this research.
Funding
This research was supported by Arak University (Grant No.: 98/11402).
Authors contributions
Proposing the model and developing the proposed model, drafting and editing the manuscript, drafting and editing the figures: Farhang Jaryani; Initiating and coordinating the project, interpreting the data, editing the manuscript, and editing the figures: Maryam Amiri.
Conflict of interest
The authors declared no conflict of interest.
Acknowledgements
We thank Iran’s National Elites Foundation and Arak University for their support in funding this research.
References
- Figueiredo DR, Azeiteiro UM, Esteves SM. Gonçalves FJM, Pereira MJ Microcystin-producing blooms-a serious global public health issue. Ecotoxicology and Environmental Safety. 2004; 59(2):151-63. [DOI:10.1016/j.ecoenv.2004.04.006] [PMID]
- Arthur M. Institute for health metrics and evaluation. Nursing Standard. 2014; 28(42):32. [DOI:10.7748/ns.28.42.32.s33]
- Akram M, Iqbal M, Daniyal M, Khan AU. Awareness and current knowledge of breast cancer. Biological research. 2017; 50(1):33. [DOI:10.1186/s40659-017-0140-9] [PMID] [PMCID]
- Bolhasani H, Amjadi E, Tabatabaeian M, Jassbi SJ. A histopathological image dataset for grading breast invasive ductal carcinomas. Informatics in Medicine Unlocked. 2020; 19:100341. [DOI:10.1016/j.imu.2020.100341]
- Veta M, Van Diest PJ, Kornegoor R, Huisman A, Viergever MA, Pluim JP. Automatic nuclei segmentation in H&E stained breast cancer histopathology images. PloS one. 2013; 8(7):e70221. [DOI:10.1371/journal.pone.0070221] [PMID] [PMCID]
- Rosen PP. Rosen’s breast pathology. Philadelphia: Lippincott Williams & Wilkins; 2001. [Link]
- Schwartz AM, Henson DE, Chen D, Rajamarthandan S. Histologic grade remains a prognostic factor for breast cancer regardless of the number of positive lymph nodes and tumor size: a study of 161 708 cases of breast cancer from the SEER Program. Archives of Pathology and Laboratory Medicine. 2014; 138(8):1048-52. [DOI:10.5858/arpa.2013-0435-OA] [PMID]
- Fergus R, Perona P, Zisserman A. Object class recognition by unsupervised scale-invariant learning. Paper presented at: 2003 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, 2003. Proceedings. 18-20 June 2003; Madison, WI, USA. [doi:10.1109/CVPR.2003.1211479]
- Netzer Y, Wang T, Coates A, Bissacco A, Wu B, Ng AY. Reading digits in natural images with unsupervised feature learning. NIPS Workshop on Deep Learning and Unsupervised Feature Learning. 2011. [Link]
- Nourmohammadi-Khiarak J, Feizi-Derakhshi MR, Razeghi F, Mazaheri S, Zamani-Harghalani Y, Moosavi-Tayebi R. New hybrid method for feature selection and classification using meta-heuristic algorithm in credit risk assessment. Iran Journal of Computer Science. 2020; 3:1-11. [DOI:10.1007/s42044-019-00038-x]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015; 521(7553):436-44. [DOI:10.1038/nature14539] [PMID]
- Turati F, Dalmartello M, Bravi F, Serraino D, Augustin L, Giacosa A, et al. Adherence to the world cancer research fund/american institute for cancer research recommendations and the risk of breast cancer. Nutrients. 2020; 12(3):607.[DOI:10.3390/nu12030607] [PMID] [PMCID]
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Paper presented at: 3rd International Conference on Learning Representations. 2015:1-14. [Link]
- Xie S, Girshick R, Dollár P, Tu Z, He K. Aggregated residual transformations for deep neural networks. Paper presented at: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 21-26 July 2017; Honolulu, HI, USA. [DOI:10.1109/CVPR.2017.634]
- Nahid AA, Kong Y. Involvement of machine learning for breast cancer image classification: A survey. Computational and Mathematical Methods in Medicine. 2017; 2017:3781951.[DOI:10.1155/2017/3781951] [PMID] [PMCID]
- Ghatak A. Convolutional Neural Networks (ConvNets). Deep learning with R. 1th ed. Singapore: Springer; 2019. [DOI:10.1007/978-981-13-5850-0_7]
- Zohrevandi P, Jaryani F. Proposing an effective framework for hybrid clustering on heterogeneous data in distributed systems. International Journal of Advanced Computer Science and Information Technology. 2018; 7(4):71-81. [Link]
- Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, inception-resnet and the impact of residual connections on learning. Proceedings of the AAAI Conference on Artificial Intelligence. 2017; 31(1):4278-84 [DOI:10.1609/aaai.v31i1.11231]
- Lu D, Weng Q. A survey of image classification methods and techniques for improving classification performance. International Journal of Remote Sensing. 2007; 28(5):823-70.[DOI:10.1080/01431160600746456]
- Olivas ES, Guerrero JDM, Martinez-Sober M, Magdalena-Benedito JR, Serrano L. Handbook of research on machine learning applications and trends: Algorithms, methods, and techniques. United States: IGI Global; 2009. [DOI:10.4018/978-1-60566-766-9]
- Nourmohammadi-Khiarak J, Mazaheri S, Moosavi-Tayebi R, Noorbakhsh-Devlagh H. Object detection utilizing modified auto encoder and convolutional neural networks. Paper presented at: 2018 Signal Processing: Algorithms, Architectures, Arrangements, and Applications (SPA). 19-21 September 2018; Poznan, Poland. [DOI:10.23919/SPA.2018.8563423]
- Spanhol FA, Oliveira LS, Petitjean C, Heutte L. A dataset for breast cancer histopathological image classification. Ieee Transactions on Biomedical Engineering. 2015; 63(7):1455-62. [DOI:10.1109/TBME.2015.2496264] [PMID]
- George YM, Zayed HH, Roushdy MI, Elbagoury BM. Remote computer-aided breast cancer detection and diagnosis system based on cytological images. IEEE Systems Journal. 2013; 8(3):949-64. [DOI:10.1109/JSYST.2013.2279415]
- Kowal M, Filipczuk P, Obuchowicz A, Korbicz J, Monczak R. Computer-aided diagnosis of breast cancer based on fine needle biopsy microscopic images. Computers in Biology and Medicine. 2013; 43(10):1563-72. [DOI:10.1016/j.compbiomed.2013.08.003] [PMID]
- Hu Q, Whitney HM, Giger ML. A deep learning methodology for improved breast cancer diagnosis using multiparametric MRI. Scientific reports. 2020; 10(1):10536. [DOI:10.1038/s41598-020-67441-4] [PMID] [PMCID]
- Araújo T, Aresta G, Castro E, Rouco J, Aguiar P, Eloy C, et al. Classification of breast cancer histology images using convolutional neural networks. PloS one. 2017; 12(6):e0177544. [DOI:10.1371/journal.pone.0177544] [PMID] [PMCID]
- Han Z, Wei B, Zheng Y, Yin Y, Li K, Li S. Breast cancer multi-classification from histopathological images with structured deep learning model. Scientific Reports. 2017; 7(1):4172.[DOI:10.1038/s41598-017-04075-z] [PMID] [PMCID]
- Ginsburg O, Yip CH, Brooks A, Cabanes A, Caleffi M, Dunstan Yataco JA, et al. Breast cancer early detection: A phased approach to implementation. Cancer. 2020; 126(S 10):2379-93. [DOI:10.1002/cncr.32887] [PMID] [PMCID]
- Faisal MI, Bashir S, Khan ZS, Khan FH. An evaluation of machine learning classifiers and ensembles for early stage prediction of lung cancer. IPaper presented at: 3rd International Conference on Emerging Trends in Engineering, Sciences and Technology (ICEEST). 21 December 2018; Karachi, Pakistan. [DOI:10.1109/ICEEST.2018.8643311]
- Bhowal P, Sen S, Velasquez JD, Sarkar R. Fuzzy ensemble of deep learning models using choquet fuzzy integral, coalition game and information theory for breast cancer histology classification. Expert Systems with Applications. 2022; 190:116167.[DOI:10.1016/j.eswa.2021.116167]
- Redmon J, Divvala S, Girshick R, Farhadi A. You only look once: Unified, real-time object detection. Paper presented at: Proceedings of the IEEE conference on computer vision and pattern recognition. July 1 2016; Las Vegas, NV, USA. [DOI:10.1109/CVPR.2016.91]
Type of Study: Original Article |
Subject:
Biostatistics
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |